MapMan Crack Full Version Download For PC [March-2022]
- graphoutatarkruser
- May 19, 2022
- 3 min read
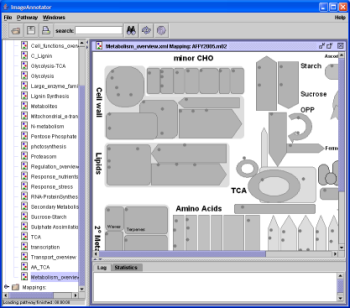
MapMan Crack Download [Mac/Win] (Updated 2022) The software is designed for the analysis of transcript expression data generated using oligonucleotide microarrays and the generation of metabolic pathways diagrams from this information. MapMan is designed for use with the Arabidopsis genome as a model, but the general framework can be adapted to other organisms for ease of use. MapMan is a visualization tool that allows the user to view transcript expression data and the metabolic processes within which these data are processed. MapMan displays information in a process-centric manner that helps the user to find relevant pathways or sub-pathways that are significantly affected by the experimental treatments applied. A central component of MapMan is the genome-wide annotation of metabolic pathways which has been described in detail in (Thimm et al. 1999). MapMan also has the capability of allowing the user to identify and compare a wide variety of other experimental parameters such as: genes that are up-regulated, genes that are down-regulated, proteins that are up-regulated, proteins that are down-regulated, proteins that contain a phosphorylated residue and protein phosphorylation site prediction tools. Uses: MapMan has been designed to facilitate the study of transcript expression data that are derived from microarray hybridisation experiments. Users of MapMan include scientists who wish to analyse these data in order to find genes that are up- or down-regulated in response to experimental treatments, and scientists who wish to analyse expression data in order to understand metabolic pathways and understand how these pathways are affected by experimental treatments. Are metabolic pathways regulated by, or affecting, DNA methylation? The occurrence of hypomethylation and hypermethylation patterns and an association between these events and seed development have previously been described (Barros et al. 2001, Meyer et al. 1999). In this study we showed that the methylation patterns of the plant genes, METHYLTRANSFERASE1 (MET1), SPUTTER1 (SPT1) and DROUGHT RESPONSE LEUCINE-BINDING PROTEIN (DRLP), in the early embryo were correlated with seed development and with the number of ‘de novo’ methylation events. We also found that the most important factor affecting the demethylation process in Arabidopsis embryos was the type of preimplantation embryonic cell, a finding that confirms a previous report (Rousseau et al. 2001). The results obtained by our study with WT Arabidopsis embryos demonstrate that hypermethylation patterns exist in MapMan Crack [Latest] 2022 MapMan is a user-driven tool that displays large datasets (e.g. gene expression data from Arabidopsis Affymetrix arrays) onto diagrams of metabolic pathways or other processes. The MapMan user-driven model allows users to interactively create pathways for analysis of transcriptomic data. MapMan is accessible via the web and offers a variety of options for pathway mapping (e.g. browse, plot, export, upload, search). Other features are multiple views of pathway diagrams, tools for functional categorization of genes, and interactive pathway map annotation. MAPMAN is designed to provide a pathway browser, so you can quickly see all the available information in your dataset. The MapMan Editor is an interactive online tool for pathway mapping, supporting both static and dynamic visualizations of pathway models. Notes: Data can be sent by FTP to the MapMan editor, but it is highly recommended that all new users upload their data using MapMan’s web interface. For this reason the current MapMan Wiki is hosted on a version of SGN’s Wiki which is restricted to registered users. Instructions: Download the MapMan database go to the download page and click on the appropriate archive button, depending on your operating system On Linux: go to the bottom of the page and select a Debian package archive (.deb) On Windows: go to the bottom of the page and select a Windows installer (i.e. setup.exe or setup.msi) On Mac OSX: go to the bottom of the page and select a.dmg file Start the MapMan software MapMan is available from Note that downloading the newest version of MapMan from this source requires downloading and installing some Python packages (e.g. python-setuptools) which are not available from default repositories, so you may need to use the instructions above. Go to the MapMan Wiki for details on how to upload and use your own data. Knowledge Base: Download and install the latest version of MapMan (v3.5): Linux: Windows: Go to the main page of the MapMan Wiki for more help: Data can be uploaded to the MapMan editor via: 1a423ce670 MapMan Crack What's New In? System Requirements For MapMan: Minimum: OS: Windows XP SP2 Processor: x86 compatible processor, 64-bit processor required Memory: 1 GB RAM Graphics: DirectX 11 compatible video adapter with 512 MB graphics RAM Hard Disk Space: 800 MB available hard disk space DirectX: Version 9.0c or higher Sound: DirectX compatible sound card Network: Broadband Internet connection Additional Notes: System Requirements: OS: Windows XP
Related links:




Comments